Breast Cancer Wisconsin Dataset - Classification
- Pratibha

- Nov 1, 2021
- 3 min read

Description :
This dataset provides information about the features which are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. These features describe characteristics of the cell nuclei present in the image. In the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: "Robust Linear Programming Discrimination of Two Linearly Inseparable Sets", Optimization Methods and Software 1, 1992, 23-34].
Recommended Model :
Algorithms to be used: random forest, svm, logistic regression etc
Recommended Project :
Prediction of developing Breast Cancer
Dataset link:
https://www.kaggle.com/uciml/breast-cancer-wisconsin-data
Overview of data
Detailed overview of dataset:
- Rows = 569
- Columns= 31
1) ID number
2) Diagnosis (M = malignant, B = benign)
Ten real-valued features are computed for each cell nucleus: a) radius (mean of distances from center to points on the perimeter) b) texture (standard deviation of gray-scale values) c) perimeter d) area e) smoothness (local variation in radius lengths) f) compactness (perimeter^2 / area - 1.0) g) concavity (severity of concave portions of the contour) h) concave points (number of concave portions of the contour) i) symmetry j) fractal dimension ("coastline approximation" - 1)
EDA [CODE]
import pandas as pd
# load data
data = pd.read_csv('data.csv')
data.head()
# check details of the dataframe
data.info()
# check the no.of missing values in each column
data.isna().sum()
# statistical information about the dataset
data.describe()
# data distribution
import seaborn as sns
import matplotlib.pyplot as plt
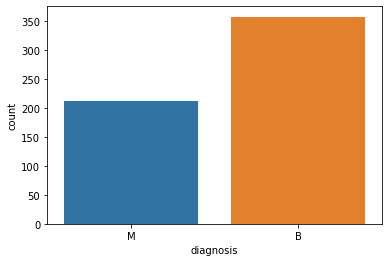
sns.countplot(x='diagnosis', data=data)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'radius_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'radius_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'radius_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'perimeter_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'perimeter_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'perimeter_worst', bins=30, kde=False)
plt.show()
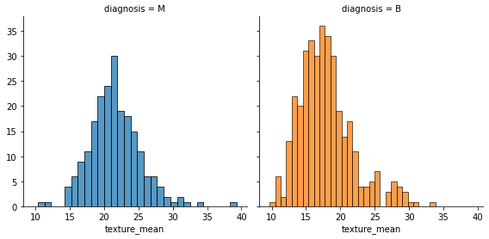
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'texture_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'texture_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'texture_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'area_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'area_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'area_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'smoothness_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'smoothness_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'smoothness_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'compactness_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'compactness_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'compactness_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'concavity_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'concavity_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'concavity_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'symmetry_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'symmetry_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'symmetry_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'concave points_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'concave points_se', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'concave points_worst', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'fractal_dimension_mean', bins=30, kde=False)
plt.show()
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'fractal_dimension_se', bins=30, kde=False)
plt.show()
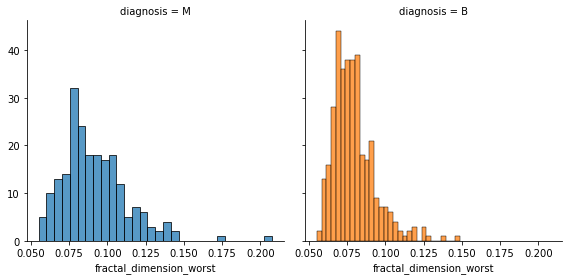
fig = sns.FacetGrid(data, col='diagnosis', hue='diagnosis', height=4)
fig.map(sns.histplot, 'fractal_dimension_worst', bins=30, kde=False)
plt.show()
Other datasets for classification:
Spam Text Dataset,
Pima Indians Diabetes Dataset
If you need implementation for any of the topics mentioned above or assignment help on any of its variants, feel free to contact us.































































Comments