Medical Cost Personal Dataset - Regression
- Pratibha
- Nov 2, 2021
- 2 min read
Updated: Nov 3, 2021

Description :
This dataset explains the cost of a small sample of USA population Medical Insurance Cost based on some attributes such as age, sex, bmi etc.
This dataset corresponds to a practical used in the book Machine Learning with R by Brett Lantz; which is a book that provides an introduction to machine learning using R. All of these datasets are in the public domain but simply needed some cleaning up and recoding to match the format in the book.
Recommended Model :
Algorithms to be used: Regression, SVM, RandomForestRegressor etc.
Recommended Project :
Medical Insurance Cost Prediction
Dataset link:
Overview of data
Detailed overview of dataset:
- Rows = 1338
- Columns= 7
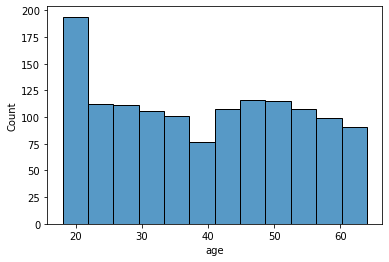
age: age of primary beneficiary
sex: insurance contractor gender, female, male
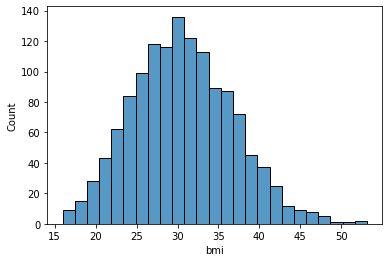
bmi: Body mass index, providing an understanding of body, weights that are relatively high or low relative to height, objective index of body weight (kg / m ^ 2) using the ratio of height to weight, ideally 18.5 to 24.9
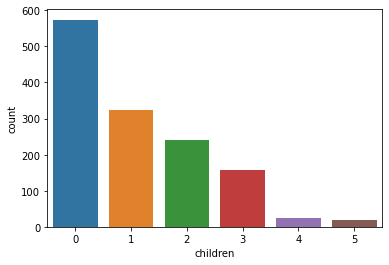
children: Number of children covered by health insurance / Number of dependents
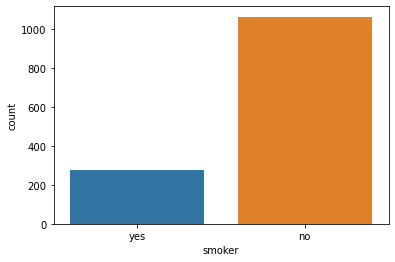
smoker: Smoking
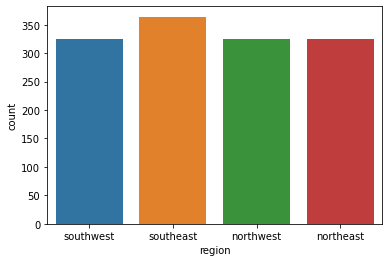
region: the beneficiary's residential area in the US, northeast, southeast, southwest, northwest.
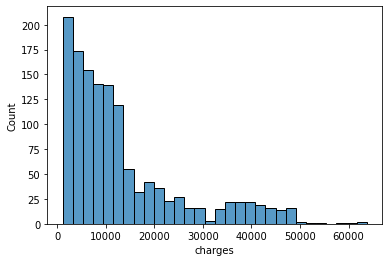
charges: Individual medical costs billed by health insurance
EDA [CODE]
import pandas as pd
# load data data = pd.read_csv('insurance.csv')
data.head()
# check details of the dataframe
data.info()
# check the no.of missing values in each column
data.isna().sum()
# statistical information about the dataset
data.describe()
# data distribution
import seaborn as sns
import matplotlib.pyplot as plt
sns.countplot(x='sex', data=data)
plt.show()
sns.countplot(x='children', data=data)
plt.show()
sns.countplot(x='smoker', data=data)
plt.show()
sns.countplot(x='region', data=data)
plt.show()
sns.histplot(data['age'], kde=False)
plt.show()
sns.histplot(data['bmi'], kde=False)
plt.show()
sns.histplot(data['charges'], kde=False)
plt.show()
Other datasets for classification:
If you need implementation for any of the topics mentioned above or assignment help on any of its variants, feel free to contact us









Comentários